import pandas as pd
df = pd.read_csv17 Yield prediction
0. Relevant packages
RDChiral
RDChiral is a wrapper for RDKit’s functionalities for reaction handling, that improves stereochemistry handling. This package will allow us to extract reaction templates from a reaction dataset, which are a standard way of encoding transformation rules.
RDChiral then also lets us apply the reaction template to a target molecule, to discover the reactants that will afford the target molecule under the given transformation.
1. Obtaining the atom mapping
To obtain the atom mapping of a reaction, you can go to this site and paste your reaction SMILES. The application will then show you the mapped reaction smiles, as well as some visualization options, including:
The atom mapping of the reaction: which atoms in the reactants correspond to each atom in the products.
The attention maps: What the underlying model is computing, that is the conection between each pair of tokens.
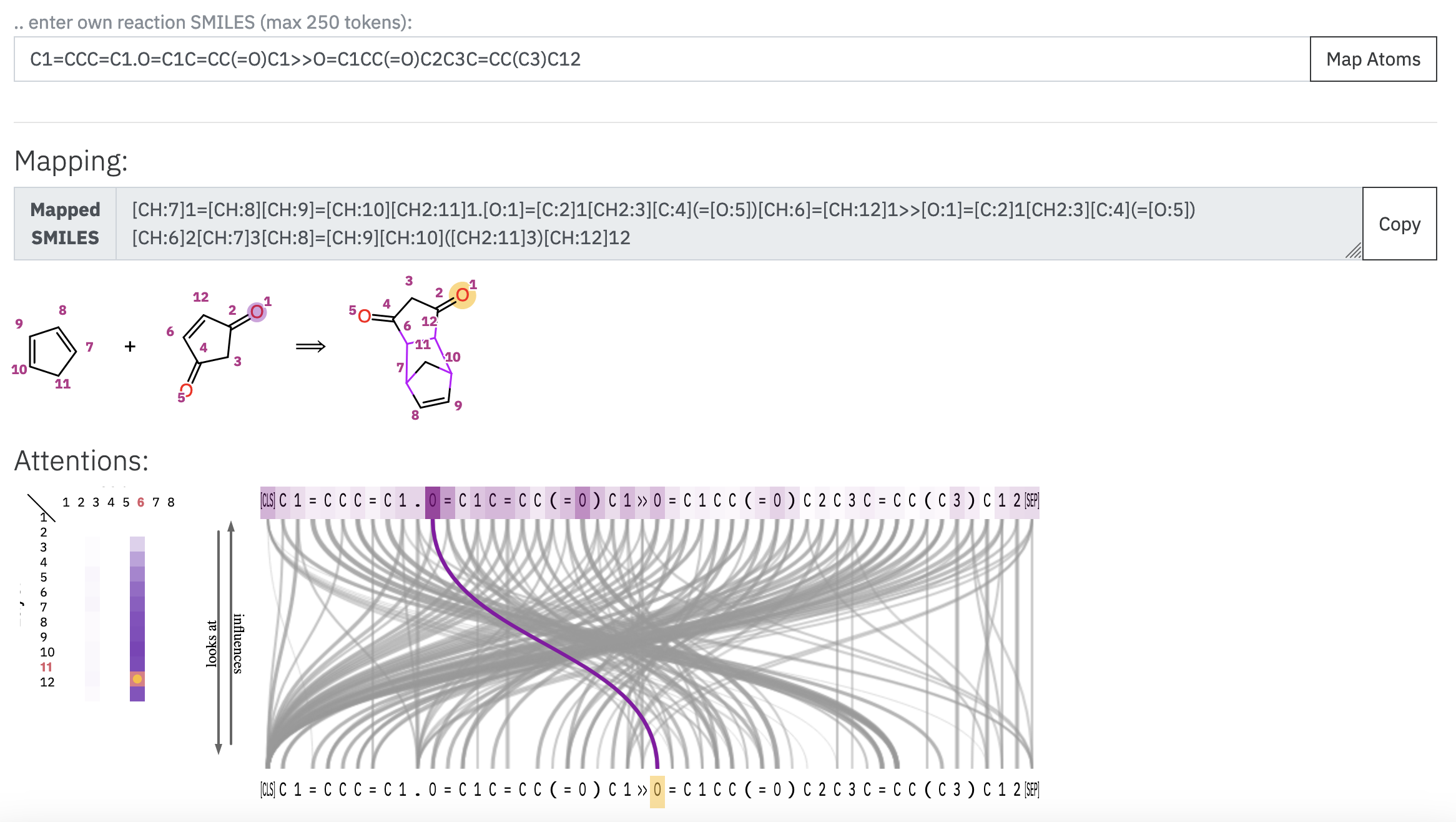
NOTE: This model is also accessible through a programming interface. For this, follow the instructions here.
TODO:
#! pip install rdkit rdchiral
! mkdir data/
! curl -L https://www.dropbox.com/sh/6ideflxcakrak10/AADN-TNZnuGjvwZYiLk7zvwra/schneider50k -o data/uspto50k.zip
! unzip data/uspto50k.zip -d data/ % Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 17 0 17 0 0 8 0 --:--:-- 0:00:02 --:--:-- 8
100 276 100 276 0 0 94 0 0:00:02 0:00:02 --:--:-- 0
0 0 0 0 0 0 0 0 --:--:-- 0:00:21 --:--:-- 0from utils import load_data, visualize_chemical_reaction
train_df, val_df, test_df = load_data()1. Reaction templates
Let’s take as an example the following coupling reaction.
rxn_example = train_df.iloc[5,0]
visualize_chemical_reaction(rxn_example)To extract the reaction template, use the extract_template function from utils.py
A reaction template describes a general transformation of some type. It describes what bonds form and break in a transformation, as well as the chemical environment of these bonds.
from utils import extract_template
tplt_example = extract_template(rxn_example)
# A reaction template looks like this
print(tplt_example)Now we can use this reaction template. Use the apply_template function from utils.py
If we use it on the same product, we should get the same reactants as above.
# Apply the extracted template to the product above.
from utils import apply_template, visualize_mols
prod_1 = rxn_example.split('>>')[1]
pred_reactants = apply_template(tplt_example, prod_1)
# This is the result of applying the template.
visualize_mols(pred_reactants[0])